Lei Su1,2, Ian P. G. Marshall2, Andreas P. Teske3, Huiqiang Yao4, Jiangtao Li1*
1 State Key Laboratory of Marine Geology, Tongji University, Shanghai 200092, China.
2 Center for Electromicrobiology (CEM), Section for Microbiology, Department of Biology, Aarhus University, Aarhus C, Denmark.
3 Department of Earth, Marine and Environmental Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
4 MLR Key Laboratory of Marine Mineral Resources, Guangzhou Marine Geological Survey, Guangzhou 510075, China.
ABSTRACT: The microbial communities of marine seep sediments contain unexplored physiological and phylogenetic diversity. Here, we examined 30 bacterial metagenome assembled genomes (MAGs) from cold seeps in the South China Sea, the Indian Ocean, the Scotian Basin, and the Gulf of Mexico, as well as from deep-sea hydrothermal sediments in the Guaymas Basin, Gulf of California. Phylogenetic analyses of these MAGs indicate that they form a distinct phylum-level bacterial lineage, which we propose as a new phylum, Candidatus Effluviviacota, in reference to its preferential occurrence at diverse seep areas. Based on tightly clustered high-quality MAGs, we propose two new genus-level candidatus taxa, Candidatus Effluvivivax and Candidatus Effluvibates. Genomic content analyses indicate that Candidatus Effluviviacota are chemoheterotrophs that harbor the Embden-eyerhof-Parnas glycolysis pathway. They gain energy by fermenting organic substrates. Additionally, they display potential capabilities for the degradation of cellulose, hemicellulose, starch, xylan, and various peptides. Extracellular anaerobic respiration appears to rely on metals as electron acceptors, with electron transfer primarily mediated by multiheme cytochromes and by a flavin-based extracellular electron transfer (EET) mechanism that involves NADH-quinone oxidoreductasedemethylmenaquinone-synthesizing enzymes, uncharacterized membrane proteins, and flavin-binding proteins, also known as the NUO-DMK-EET-FMN complex. The heterogeneity within the Ca. Effluviviacota phylum suggests varying roles in energy metabolism among different genera. While NUO-DMK-EET-FMN electron transfer has been reported predominantly in Gram-positive bacteria, it is now identified in Ca. Effluviviacota as well. We detected the presence of genes associated with bacterial microcompartments in Ca. Effluviviacota, which can promote specific metabolic processes and protect the cytosol from toxic intermediates.
Full article:https://journals.asm.org/doi/10.1128/mbio.00992-24
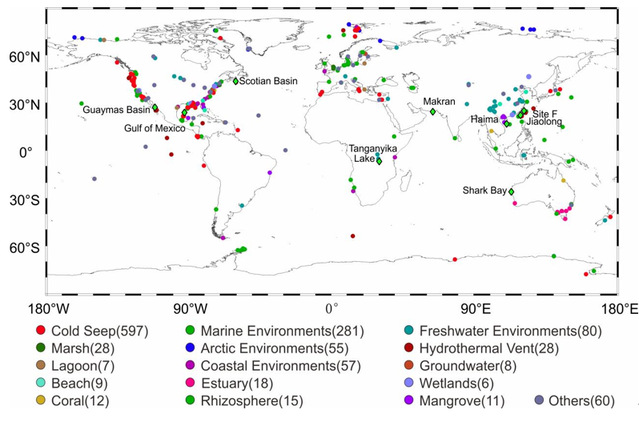
Global occurrences of CandidatusEffluviviacota genomes and 16S rRNA gene sequences.


